1.LncPCD Introduction
Programmed cell death (PCD) refers to controlled cell death that is conducted to maintain the stability of the internal environment. LncRNAs are involved in the progression of PCD in various diseases. Here, we developed LncPCD, a comprehensive database that provides information on experimentally supported associations of lncRNA-mediated PCD with diseases. The current version of LncPCD documents 6666 associations between five common types of PCD (apoptosis, autophagy, ferroptosis, necroptosis and pyroptosis) and 1222 lncRNAs in 331 diseases. We also manually curated a wealth of information: (1) 7 important lncRNA regulatory mechanisms. (2) 310 PCD-associated cell types in three species, (3) detailed information on lncRNA subcellular locations, and (4) clinical applications for lncRNA-mediated PCD in diseases. Additionally, 10 single-cell sequencing datasets were integrated into LncPCD to characterize the dynamics of lncRNAs in diseases. Overall, LncPCD represents an extremely useful resource for understanding the functions and mechanisms of lncRNA-mediated PCD in diseases.
2.Home Page
On the Home page, LncPCD provides a user-friendly searching and browsing interface. The primary features of the database are provided in menu bar form (step 1 in Figure 1). LncPCD offers the Quick Search feature, which allows users to select all or particular species types (e.g., Homo sapiens) from the species drop-down menu. After setting the species type, users can type the lncRNA of interest (e.g., HOTAIR) to query related associations in the LncPCD (step 2 in Figure 1). In the Overview section, we summarize the LncPCD with text (step 3 in Figure 1). In the Five Types of Programmed Cell Death section, we provide five clickable pictures with morphological characteristics of PCD; users can click pictures of interest to view the associations between lncRNA-mediated PCD and diseases (step 4 in Figure 1). At the bottom, the statistics of LncPCD are displayed on the left side (step 5 in Figure 1), and the world map is displayed on the right side (step 6 in Figure 1).
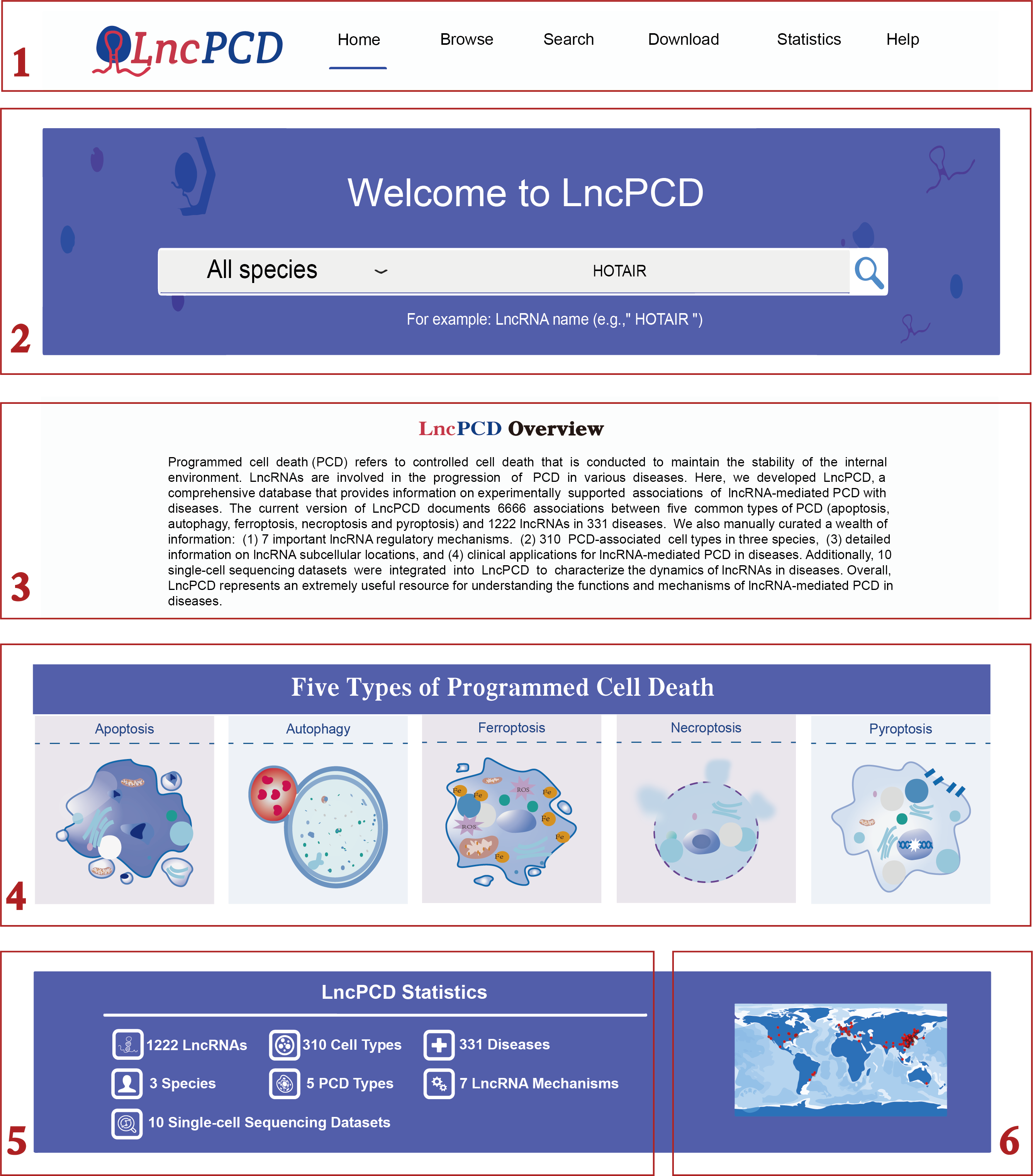
Figure 1
3. Browse Page
The Browse page was built based on a standardized classification scheme of diseases (according to the Disease Ontology database) and different hierarchical classifications. Users can browse the database by clicking the name on the left tree view. To browse the entries related to the disease of interest, users can click "Disease" and select the disease of interest (as shown in Figure 2 below, taking the disease "prostate cancer" as an example). Users can browse all entries associated with an event of interest (LncRNA Name, Species, Programmed Cell Death and LncRNA Mechanism) in a similar way. In addition, the Browse page provides statistical information on lncRNA-associated programmed cell death in diseases.
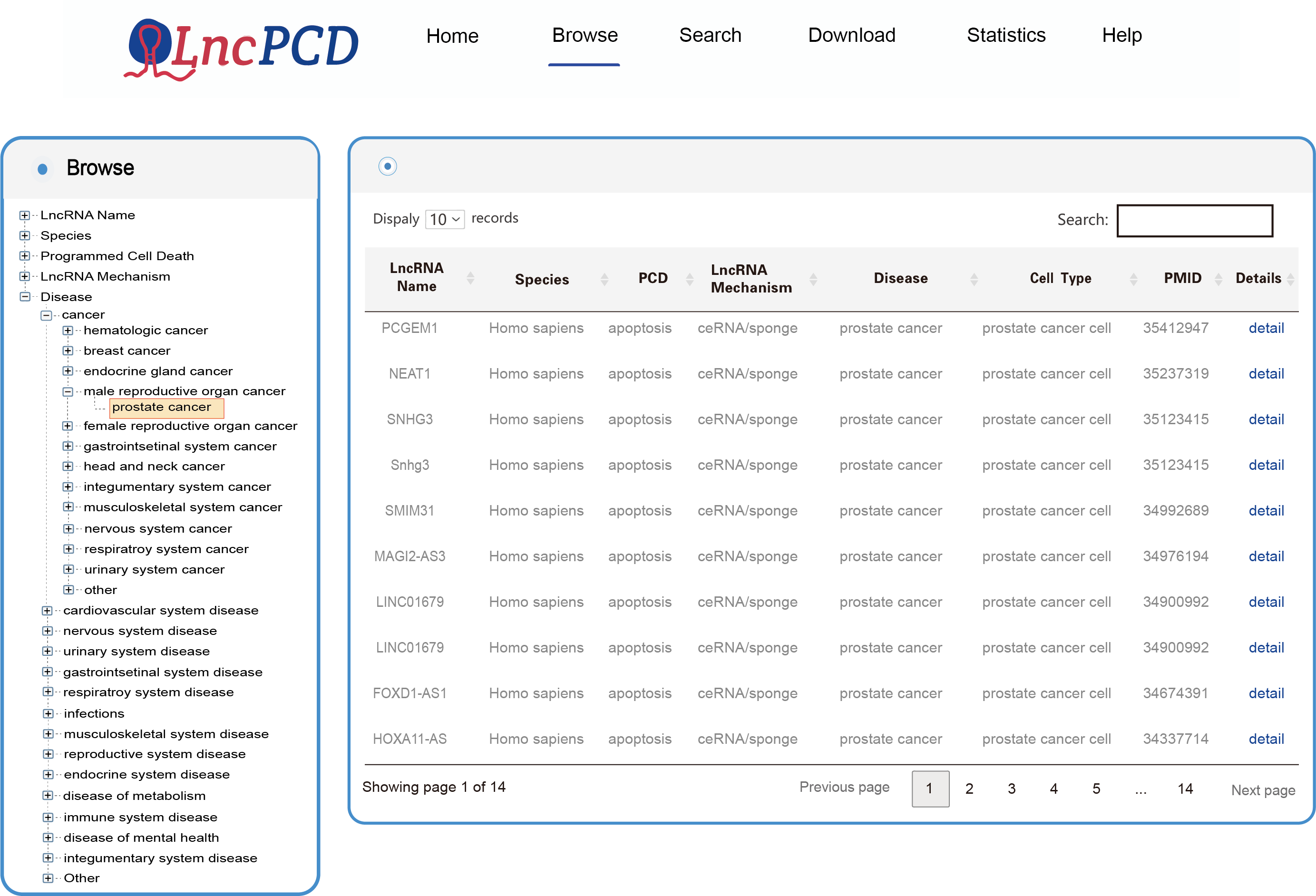
Figure 2
4. Search Page
The Search page provides two panels for searching experimentally supported associations of lncRNA-mediated PCD with diseases. The Quick Search feature allows users to select all or particular species types (e.g., Homo sapiens) from the species drop-down menu. After setting the species type, users can type the LncRNA of interest (e.g., HOTAIR) to query related associations in LncPCD (step 1 in Figure 3). The Advanced Search feature provides multiple options for a customized search of the content of interest. Users can query the associations between lncRNA-mediated PCD and diseases by combining different keywords, including the lncRNA name, species, type of PCD, lncRNA regulatory mechanism and disease (an example is shown in step 2 in Figure 3).
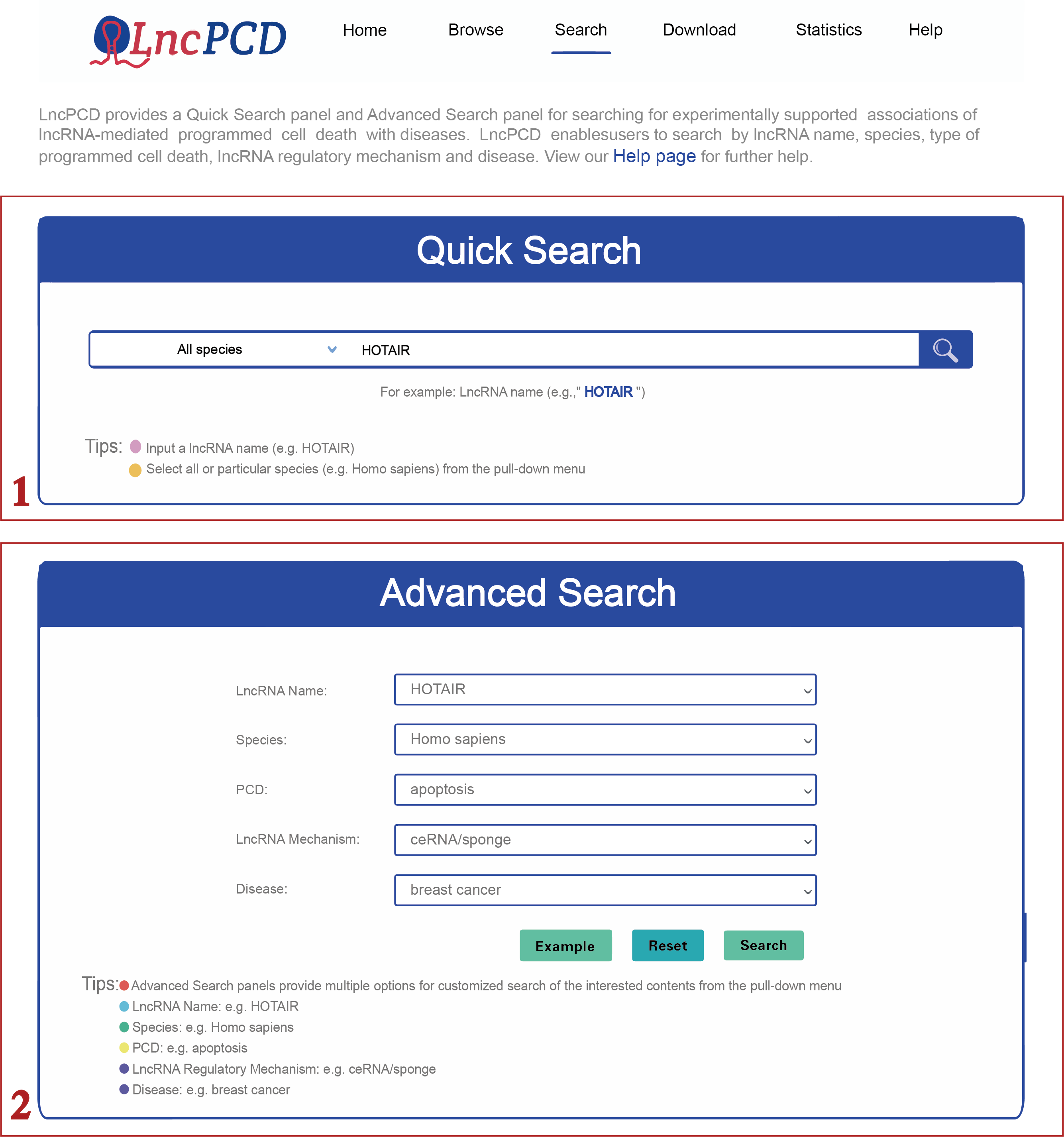
Figure 3
5. Results Table
The Search and Browse results in LncPCD are organized in a data table (as shown in Figure 4), with a single association record on each line: LncRNA name, Species, PCD, LncRNA Mechanism, Disease, Cell Type, PMID, Details (hyperlinks that can be clicked to obtain more detailed information).
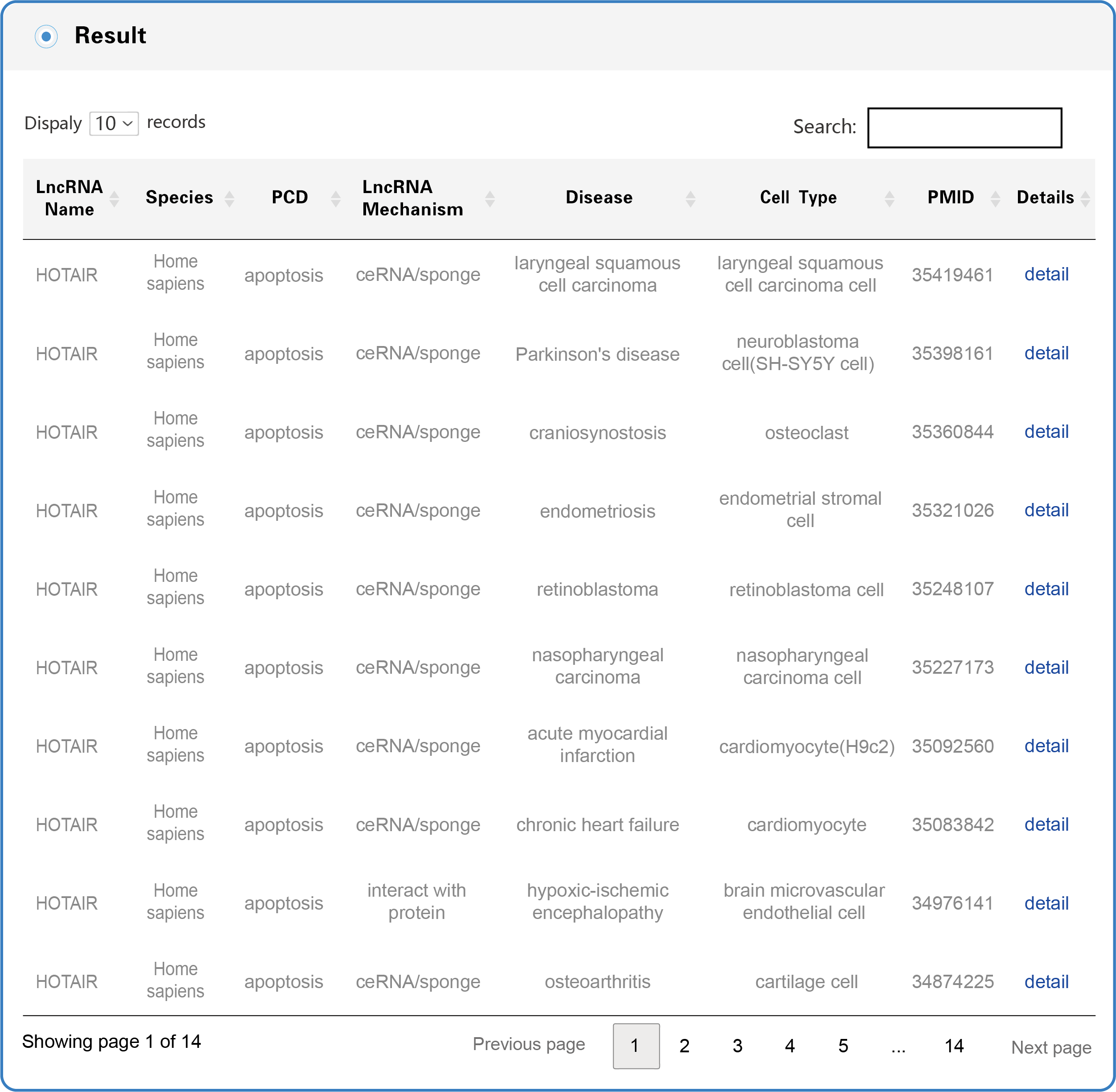
Figure 4
6. Detailed Information
To further learn about and explore the associations between lncRNA-mediated PCD and diseases, users can click "Details" in the Results Table to obtain the corresponding metadata (as shown in Figures 5-12 below).
(1) The manually curated information contains basic information and evidence, including the lncRNA name, Ensembl ID (each lncRNA Ensembl ID hyperlinks to authoritative annotation databases), species, PCD, lncRNA regulatory mechanism, disease name, cell type, expression of the lncRNA in disease, lncRNA target, cell death pathway, clinical application of lncRNA, detection method, functional description and corresponding literature (publication year, PubMed ID and publication title) (Figure 5).
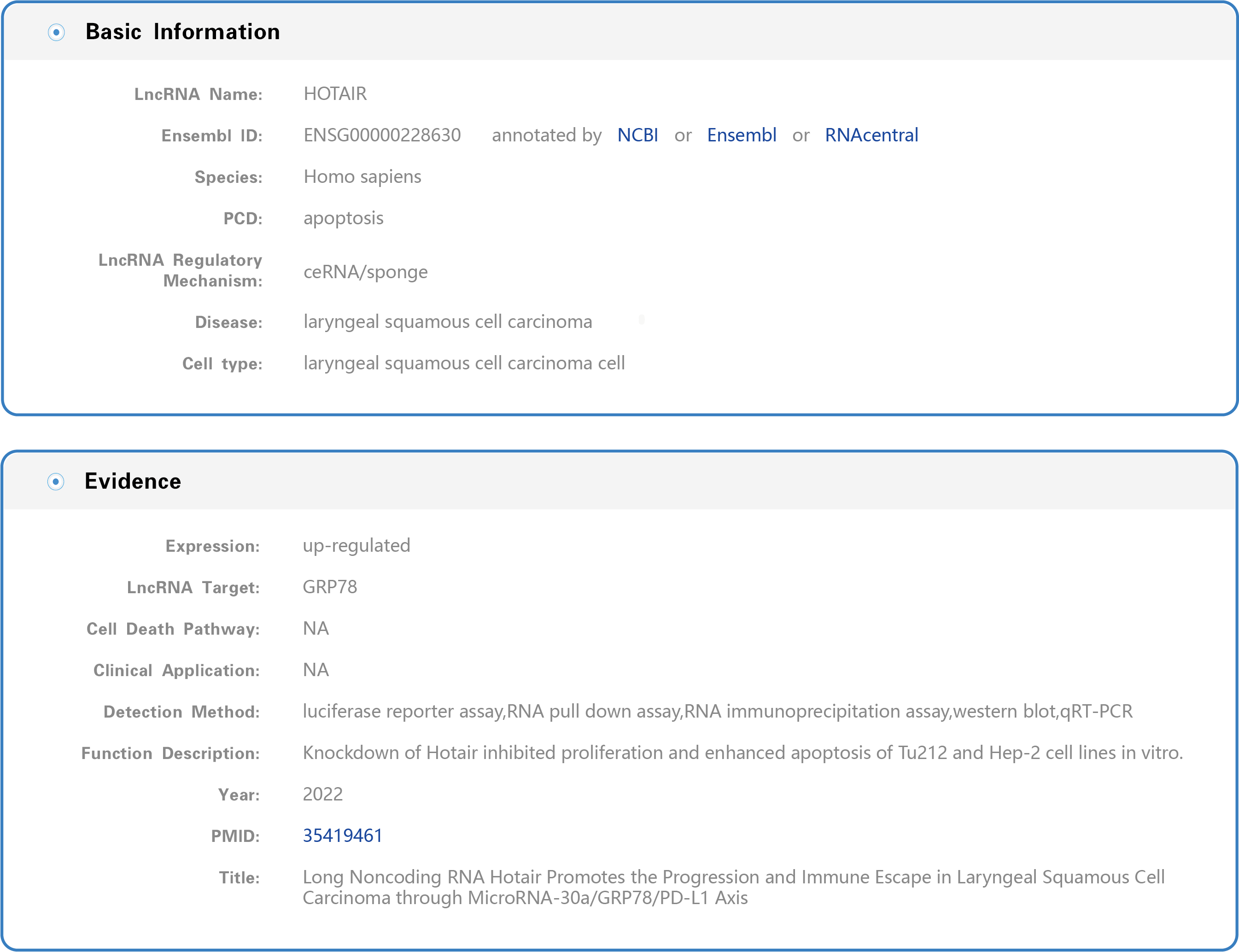
Figure 5
(2) In the cellular localization section, LncPCD provides all possible subcellular locations of the retrieved lncRNA, and the identified locations are marked with blue frames. In addition, users can obtain detailed subcellular location information by clicking on the "Detailed Subcellular Table" button (1 in Figure 6). The detailed information includes the LncRNA name, Ensembl ID, species, subcellular location, identified tissue or cell line and data source (2 in Figure 6).
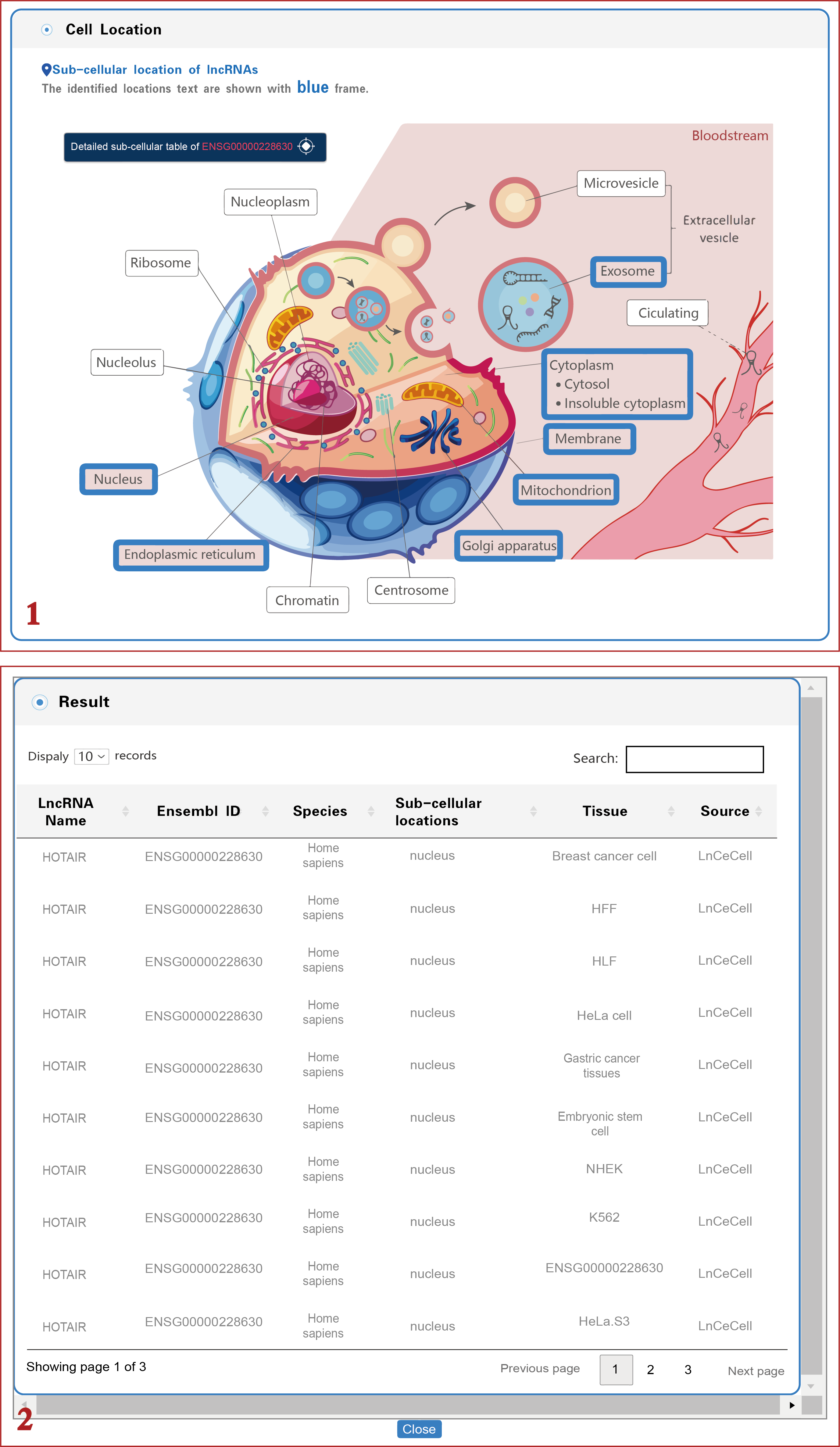
Figure 6
(3) In the section with the lncRNA cell map, LncPCD offers a scRNA-seq web tool, a user-friendly online analysis module that enables rapid and customizable visualization of PCD-associated lncRNAs, including general information presentation, clustering, cluster function, featureplot and heatmap for lncRNAs based on 10 single-cell datasets across 9 cancer types (Figure 7). Users can select a cell type (e.g., GSE77308_breast cancer cell) from the cell name drop-down menu (boxed in red in Figure 7). After setting the type of cell, users can click on the "Cluster" button, and the cluster function will generate a cluster plot from cluster analysis based on single-cell transcriptome expression data using UMAP dimensionality reduction methods (Figure 8). Users can click on the "Cluster function" button, and the cluster function will generate a plot of the GO enrichment of cluster (Figure 9) and a plot of the Pathway enrichment of the cluster (Figure 10) based on cluster analysis. Users can also click on the "Featureplot" button, and the featureplot function will generate a feature plot of the expression patterns of PCD-associated lncRNAs based on selected scRNA-seq datasets (Figure 11). In addition, users can click on the "Heatmap" button, and the heatmap function will generate a cluster analysis heatmap based on differentially expressed genes (Figure 12).
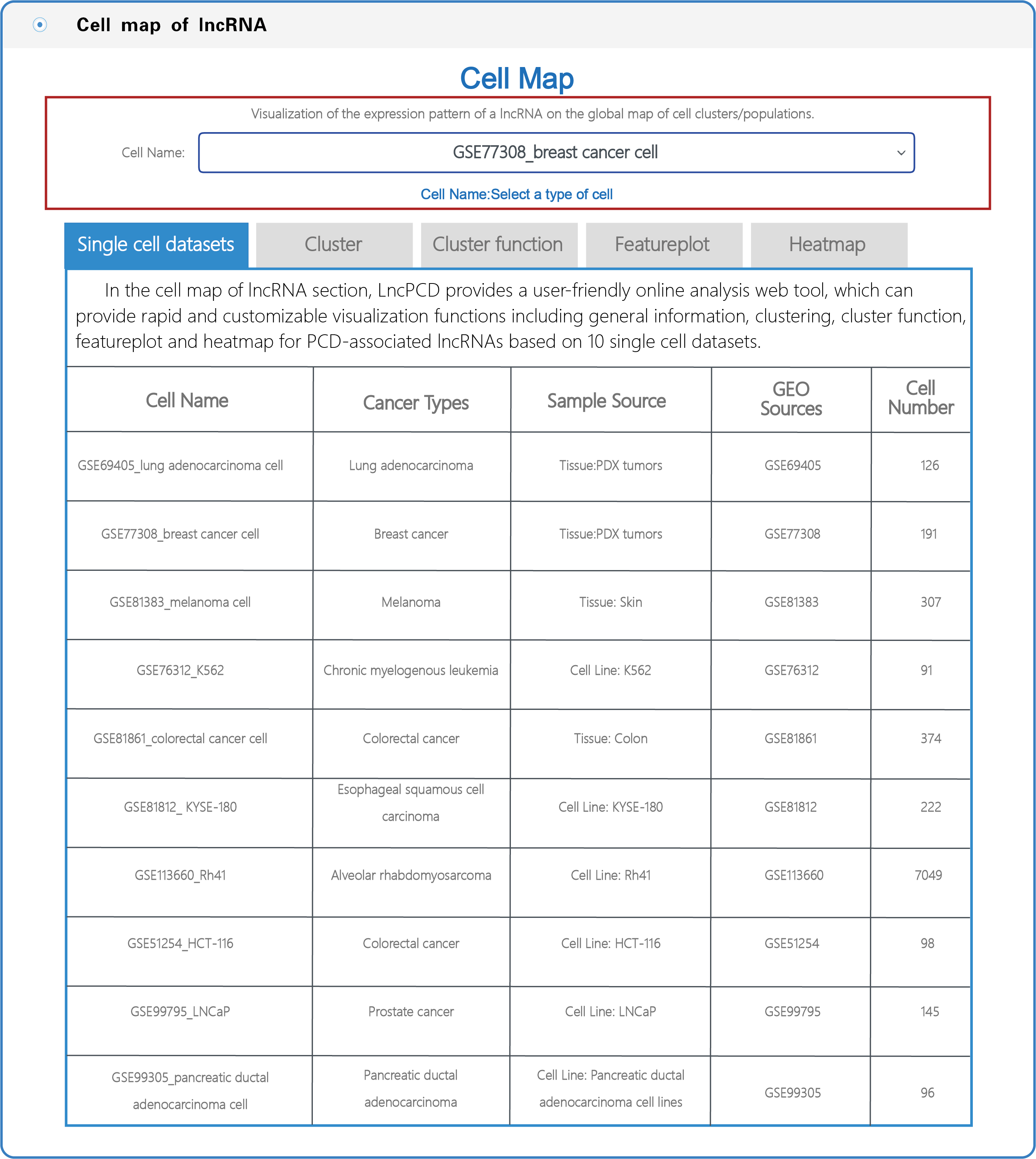
Figure 7
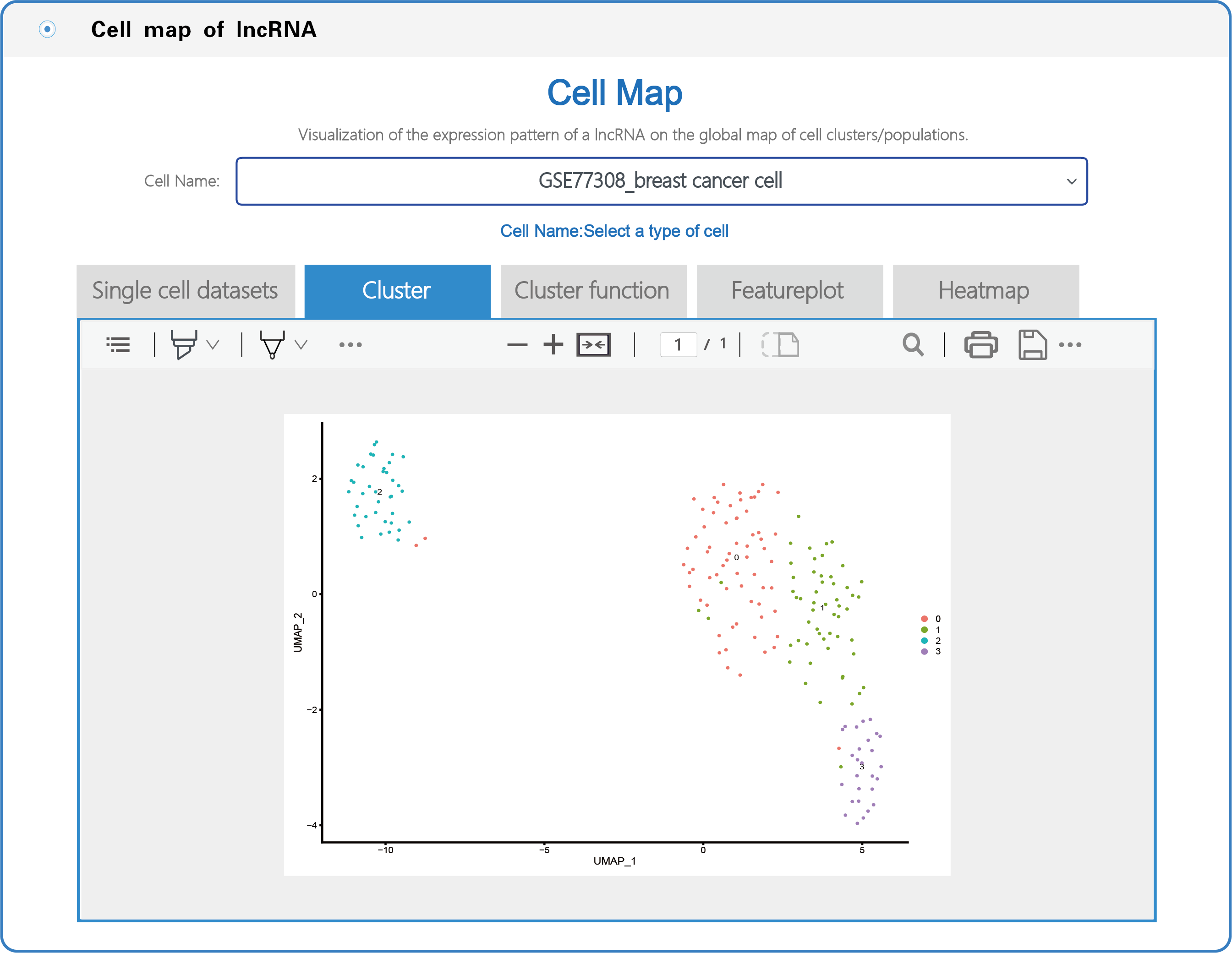
Figure 8
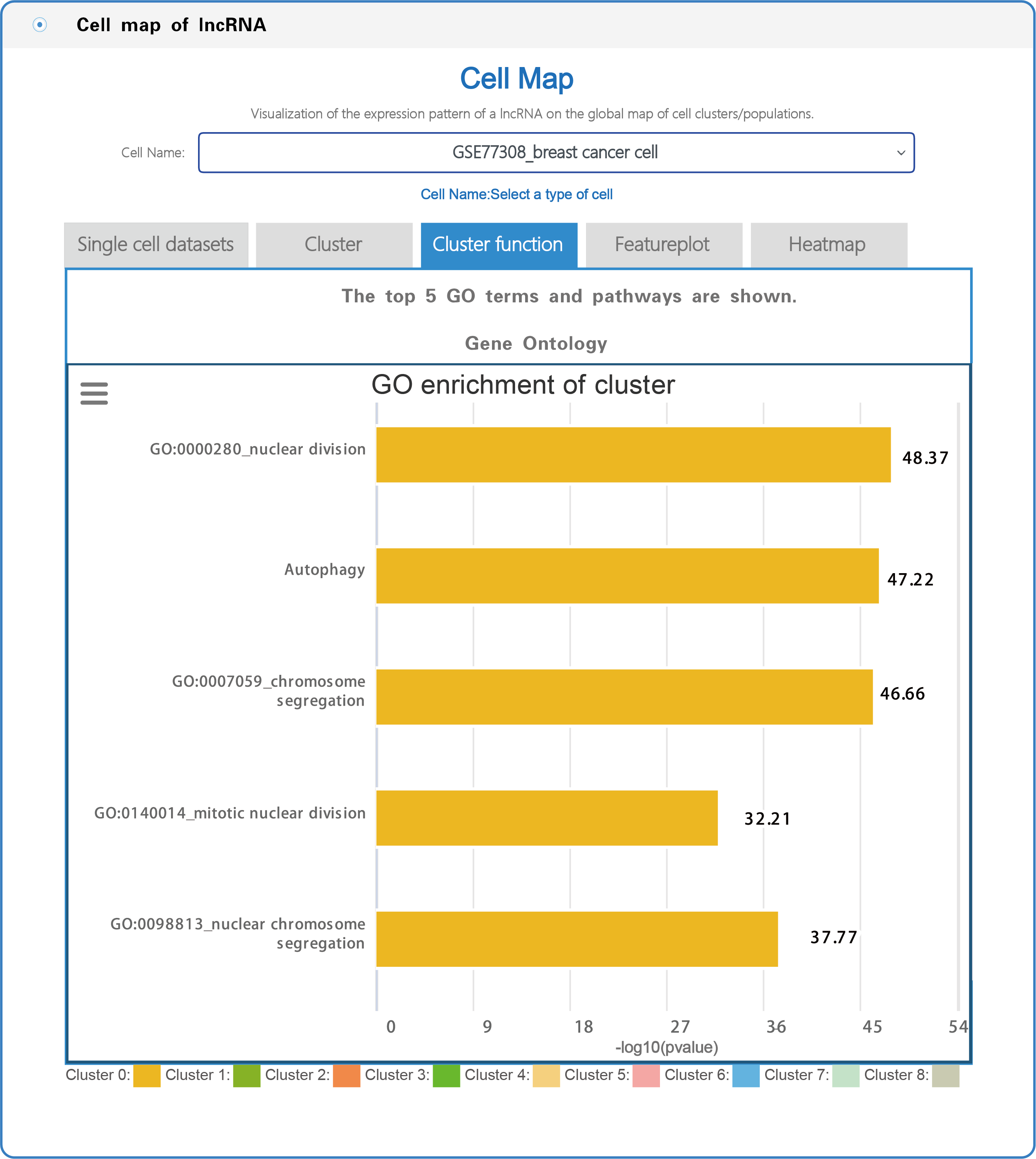
Figure 9
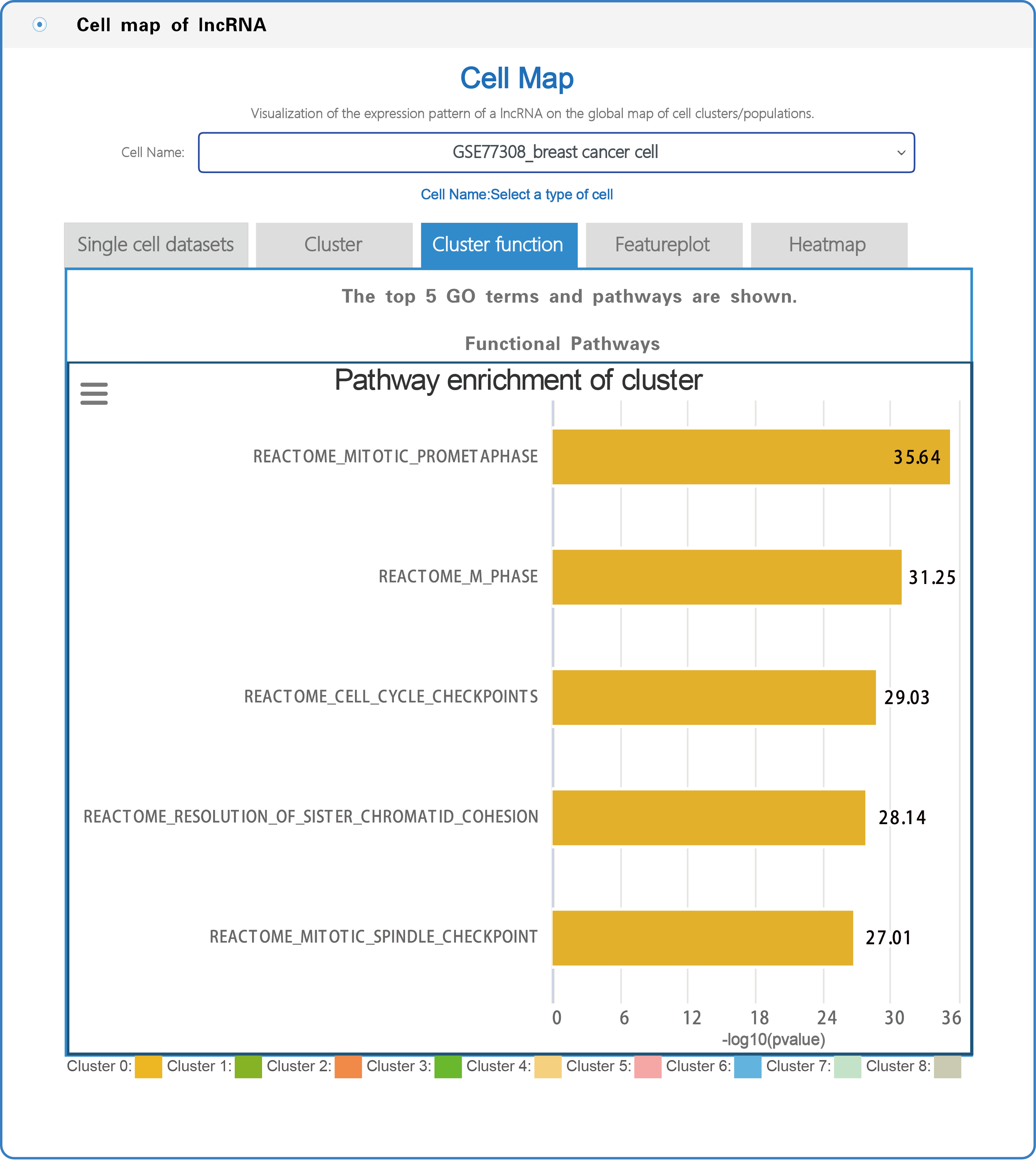
Figure 10
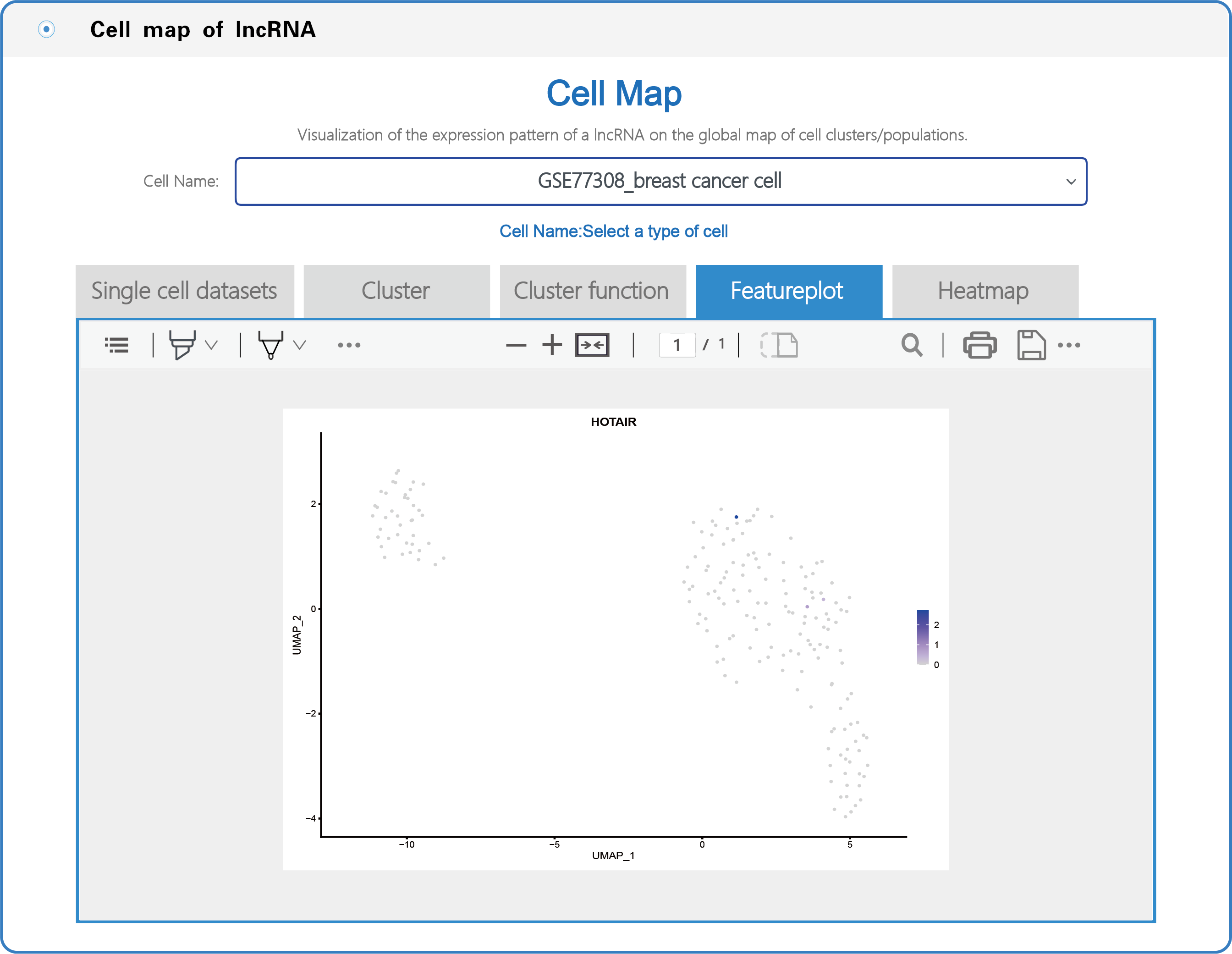
Figure 11
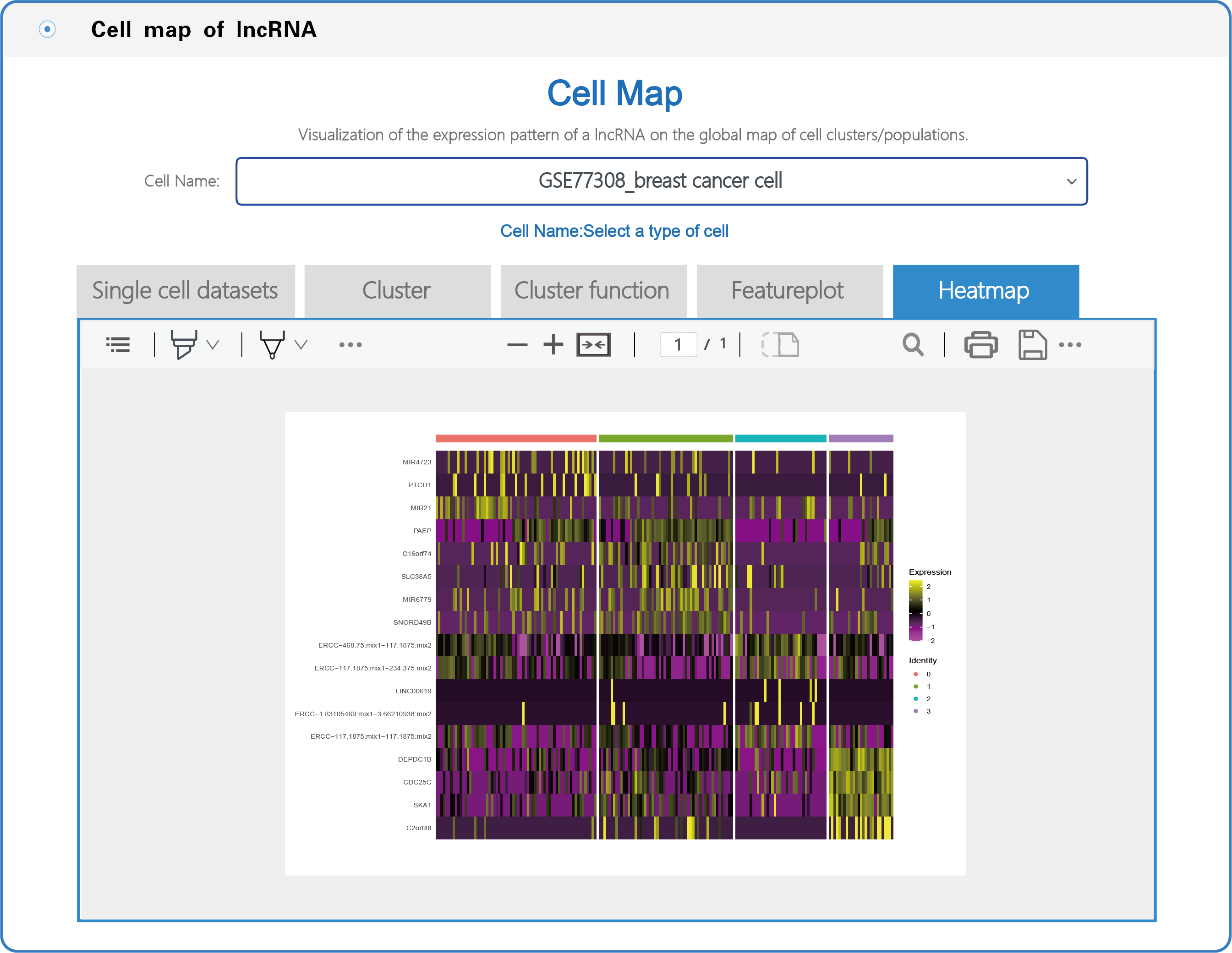
Figure 12
7. Download Data
On the download page, LncPCD provides a downloadable file in TEXT format. The data available for download include Homo sapiens-associated data, Mus musculus-associated data, Rattus norvegicus-associated data, data on the five types of PCD, lncRNA subcellular localization information, single-cell sequencing datasets, hotspot data and all data. On the left side of the page, LncPCD provides a representative visualization of all data. Users can download corresponding data by clicking on the appropriate picture. On the right side of the page, LncPCD also provides a visual map of the tag cloud that displays hotspot data. Users can download hotspot data by clicking on the appropriate area (Figure 13).
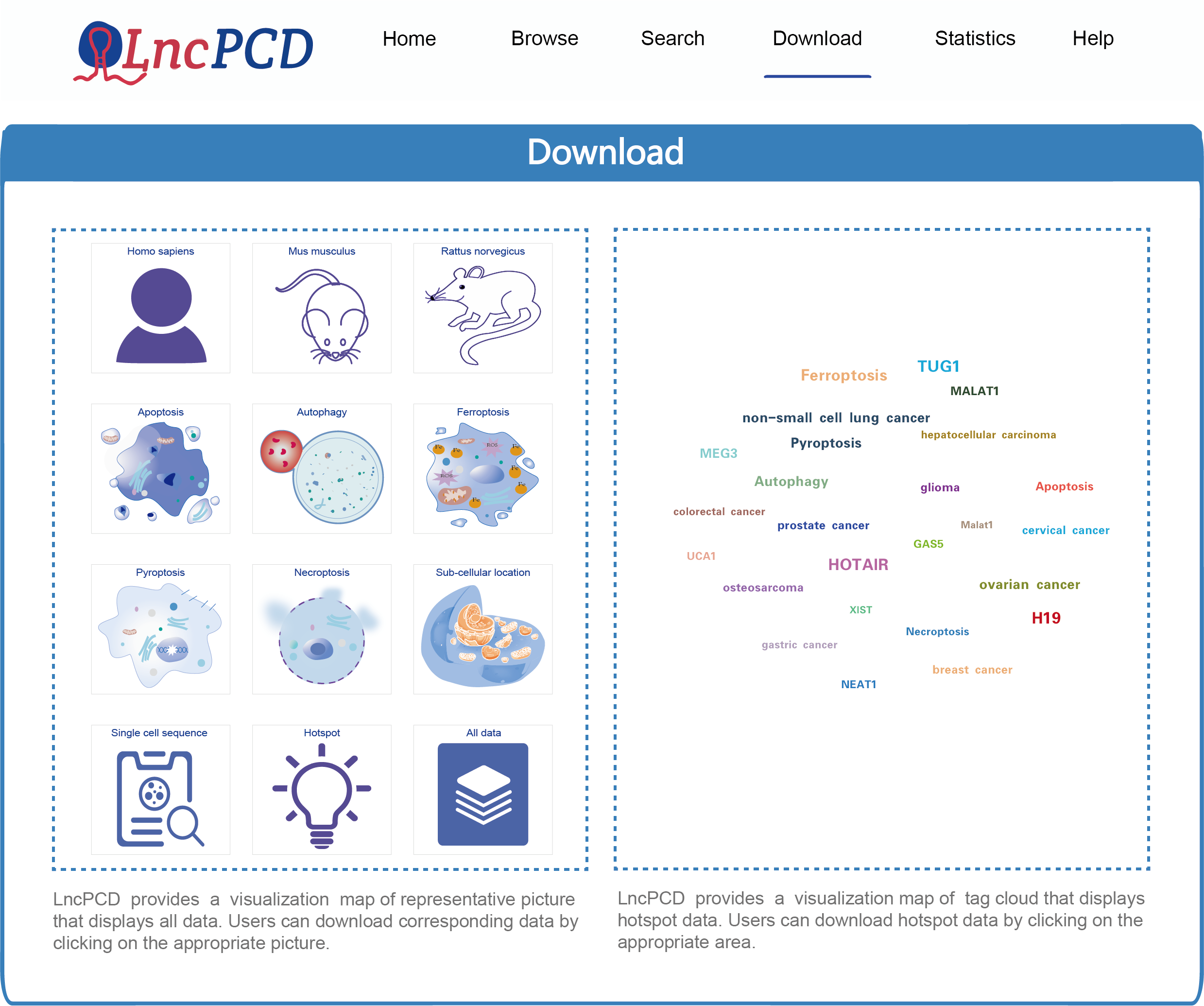
Figure 13
8. Contact
Please contact us if you have any questions using this website.
194 Xuefu Road, Harbin 150081, CHINA
wangjian_427@163.com